In a post last week I talked a bit about Permutation (Randomization) tests, and how they differ from the (classical parametric) testing procedure that we generally use in econometrics. I'm going to assume that you've read that post.
(There may be a snap quiz at some point!)
I promised that I'd provide a regression-based example. After all, the two examples that I went through in that previous post were designed to expose the fundamentals of permutation/randomization testing. They really didn't have much "econometric content".
In what follows I'll use the terms "permutation test" and "randomization test" interchangeably.
What we'll do here is to take a look at a simple regression model and see how we could use a randomization test to see if there is a linear relationship between a regressor variable, x, and the dependent variable, y. Notice that I said a "simple regression" model. That means that there's just the one regressor (apart from an intercept). Multiple regression models raise all sorts of issues for permutation tests, and we'll get to that in due course.
There are several things that we're going to see here:
- How to construct a randomization test of the hypothesis that the regression slope coefficient is zero.
- A demonstration that the permutation test is "exact". That it, its significance level is exactly what we assign it to be.
- A comparison between a permutation test and the usual t-test for this problem.
- A demonstration that the permutation test remains "exact", even when the regression model is mi-specified by fitting it through the origin.
- A comparison of the powers of the randomization test and the t-test under this model mis-specification.
The Monte Carlo experiment
A lot of this information will be revealed by means of a Monte Carlo experiment. The associated R code can be downloaded from the code page for this blog. (Even if you're not an R user, you can read the code file with any text editor, and there are lots of comments in it to help you.)
Now, let's get down to business. First, let's look at the regression model that we'll be working with. It's of the form:
yi = β1 + β2 xi + εi ; i = 1, 2, 3, ...., n. (1)
The regressor, x, is non-random, but apart from that we're not really going to make any assumptions beyond "exchangeability" of the sample observations. (See my previous post for an explanation of this.)
If (1) is the Data-Generating Process (DGP), as well as the form of the model that is estimated by OLS, then there is no model mis-specification. On the other hand if (1) is the DGP, but we "under-fit" the regression by omitting the intercept, then we have a mis-specified model. In the latter case, we know that this has adverse implications for the OLS estimator and for the conventional tests that we conduct.
You'll know by now that to conduct a permutation test we have to choose a suitable test statistic, S. Given the null hypothesis that we want to test, one option would be to choose S = b2, where b2 is the OLS estimator for β2. That would be just fine, even though it's not a pivotal statistic. Then we could keep the ordering of the y values in the sample, and consider permutations of the x data (or vice versa - it wouldn't matter). See Edgington (1987) or Noreen (1989).
Alternatively, we could recall that b2 is proportional to the (Pearson) simple correlation coefficient between y and x. In fact the proportionality "constant" is the ratio of the sample standard deviations of y and x. This ratio stays the same as we permute the sample values. Moreover, for a set of x and y values, the correlation coefficient increases monotonically with b2.
So, all we have to do is to test the null hypothesis of "no correlation", and this will serve our purpose precisely. The alternative hypothesis will be 2-sided, namely that there is a linear correlation between x and y. This is a problem that was dealt with in Example 2 of my last post on permutation tests, so we know already what's involved.
So, all we have to do is to test the null hypothesis of "no correlation", and this will serve our purpose precisely. The alternative hypothesis will be 2-sided, namely that there is a linear correlation between x and y. This is a problem that was dealt with in Example 2 of my last post on permutation tests, so we know already what's involved.
This time our R code will be a bit more efficient than before. We'll take advantage of Steve Garren's muOutlier package for R to implement the permutation test.
Sample sizes of n = 15, 30, and 60 are are considered separately. (This is hardly a situation where we can appeal to "large-sample asymptotics".) The number of permutations are 1.308x1012, 2.653x1032, and 8.321x1081 for n = 15, 30, and 60 respectively. Obviously, we'll just use a random selection of these possible permutations! For n = 15, 30 we'll use 2,000 selections; and we'll use 5,000 selections for n = 60. A bit of experimentation shows that these numbers are sufficient for the accuracy that we want, as is the number of Monte Carlo replications that we'll use, namely 2,000.
Sample sizes of n = 15, 30, and 60 are are considered separately. (This is hardly a situation where we can appeal to "large-sample asymptotics".) The number of permutations are 1.308x1012, 2.653x1032, and 8.321x1081 for n = 15, 30, and 60 respectively. Obviously, we'll just use a random selection of these possible permutations! For n = 15, 30 we'll use 2,000 selections; and we'll use 5,000 selections for n = 60. A bit of experimentation shows that these numbers are sufficient for the accuracy that we want, as is the number of Monte Carlo replications that we'll use, namely 2,000.
Within each replication of the Monte Carlo experiment what we do is construct and apply the randomization test. We compute its p-value and then keep track of those p-values that are less than or equal to our desired significance level, which will be 5%. If the null hypothesis is true, the observed fraction of such p-values over our 2,000 Monte Carlo replications will be 5%. (Any departure from this value would be due to using too few replications, or two few randomly chosen permutations.)
At each replication of the experiment we also apply the usual t-test for a zero slope. Note that the latter is based on lots of assumptions - such as errors that independent, homoskedastic, and Normally distributed - whereas the permutation test is free of these.
Significance levels
To get a "fair" comparison between the two type of tests at the outset, the DGP in (1) uses random disturbances that are i.i.d. Normal, with a zero mean and a constant variance. So, When the null hypothesis is true and the model that is estimation is correctly specified, the t-test should reject the null hypothesis in 5% of the replications of the experiment if we use the appropriate Student-t critical value(s).
So, with this in mind, let's take a look at a partial set of our results, for the case where n = 30. In Table 1, the regression is fitted through the origin, so unless the value of β1 in the DGP is zero, the estimated model would be mis-specified:
Table 1
We'll come back to this table shortly, but for now just focus on the row that is highlighted in light green. This corresponds to the case where both β1 and β2 are zero. That is, the DGP in (1) has no intercept, so the model is correctly specified, and the null hypothesis of a zero slope is true.
Because the null hypothesis is true, the power of the test is just its significance level (5%). Notice that the reported empirical significance levels match the anticipated 5%!
This is good news. Given the particular errors that were used in the DGP in the simulations, this had to happen for the t-test. However, the result for the randomization test could be misleading. Why? Well. we could obtain the 5% rejection rate correctly, even if the distribution of the p-values from which this rate was calculated is "weird".
In an old post on this blog I discussed the sampling distribution of a p-value. One point that I covered was that if the null hypothesis is true, then this sampling distribution has to be Uniform on [0 , 1], regardless of the testing problem! This result gives another way of checking if the code for our simulation experiment is performing accurately, and that we've used enough replications and random selections of the permutations.
Figure 2 shows the distribution of the 2,000 p-values for the permutation test, for the situation in the first row of results in Table 1:
As we can see, this distribution is "reasonably uniform", as required. More formally, using the uniftest package in R we find that the Kolmogorov-Smirnov test statistic for uniformity is D = 0.998 (p = 0.24); and the Kuiper test statistic is V = 1.961 (p = 0.23). So, we seem to be in good shape here.
So far, all that we seem to have shown is that the permutation test and the t-test exhibit no "size-distortion" when the model is correctly specified, and the errors satisfy the assumptions needed for the t-test to be valid. Well, whoopee!
Let's take a look back at Table 1, and now focus on the second line of results (highlighted in orange). Recall that the estimated model omits the intercept - the model is fitted through the origin. However, now, in Table 1, β1 = 1, so the DGP includes an intercept and the fitted model is under-specified. We've omitted a (constant) regressor from the estimated model.
In this case the usual t-statistic follows a non-central Student-t distribution, with a non-centrality parameter that increases monotonically with β12, and which depends on the x data and the variance of the error term. It's value is unobservable! And we can't use the critical value(s) from the non-central t distribution if we don't know the value of the non-centrality parameter.
Obviously, the usual (central) Student-t critical values are no longer correct, and the observed significance level (the rejection rate of the null in the experiment when the null is true) will differ from 5%. Depending on the situation, it may be less than 5%, or greater than 5%. The extent of this difference is the "size-distortion" associated with the test when we mis-apply it in this way.
In the second line of Table 1 we see two important things. First, the t-test has a downwards size-distortion. We wanted to apply the test at the 5% significance level, but in fact it only rejected the null, when it was true, 1% of the time! Of course, if we just mis-applied this test once, in an application, we would have no idea if there was any substantial size-distortion or not.
The second (really neat) thing that we see in the second line of that table is that the permutation test still has a significance level of 5%! Even though the model is mis-specified, this doesn't affect the test - at least in terms of it still being "exact" with respect to the significance level that we wanted to achieve.
And this is a totally general result!
Power considerations
Figure 1
As we can see, this distribution is "reasonably uniform", as required. More formally, using the uniftest package in R we find that the Kolmogorov-Smirnov test statistic for uniformity is D = 0.998 (p = 0.24); and the Kuiper test statistic is V = 1.961 (p = 0.23). So, we seem to be in good shape here.
So far, all that we seem to have shown is that the permutation test and the t-test exhibit no "size-distortion" when the model is correctly specified, and the errors satisfy the assumptions needed for the t-test to be valid. Well, whoopee!
Let's take a look back at Table 1, and now focus on the second line of results (highlighted in orange). Recall that the estimated model omits the intercept - the model is fitted through the origin. However, now, in Table 1, β1 = 1, so the DGP includes an intercept and the fitted model is under-specified. We've omitted a (constant) regressor from the estimated model.
In this case the usual t-statistic follows a non-central Student-t distribution, with a non-centrality parameter that increases monotonically with β12, and which depends on the x data and the variance of the error term. It's value is unobservable! And we can't use the critical value(s) from the non-central t distribution if we don't know the value of the non-centrality parameter.
Obviously, the usual (central) Student-t critical values are no longer correct, and the observed significance level (the rejection rate of the null in the experiment when the null is true) will differ from 5%. Depending on the situation, it may be less than 5%, or greater than 5%. The extent of this difference is the "size-distortion" associated with the test when we mis-apply it in this way.
In the second line of Table 1 we see two important things. First, the t-test has a downwards size-distortion. We wanted to apply the test at the 5% significance level, but in fact it only rejected the null, when it was true, 1% of the time! Of course, if we just mis-applied this test once, in an application, we would have no idea if there was any substantial size-distortion or not.
The second (really neat) thing that we see in the second line of that table is that the permutation test still has a significance level of 5%! Even though the model is mis-specified, this doesn't affect the test - at least in terms of it still being "exact" with respect to the significance level that we wanted to achieve.
And this is a totally general result!
Power considerations
So much for the significance level, and size-distortion. What about the powers of the tests that we're considering?
If we move down Table 1 we still have a mis-specified model (because β1 ≠ 0), but the value of the slope coefficient is increasing. That is, the null hypothesis is becoming more and more false. We're now looking at the power functions for the tests. Because the size of the t-test is distorted downwards, it's not surprising that this test has lower power than the permutation test in this particular case. (More generally, that need not be the case - the power curves could intersect at some point.)
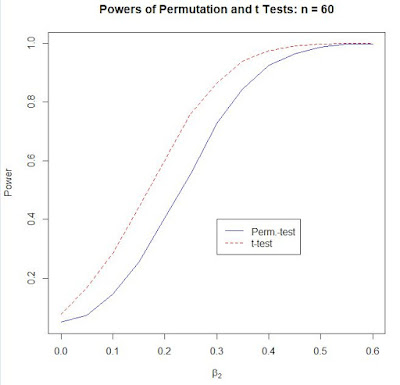
Figures 2 and 3 show the corresponding power curves when n = 15, and n = 60. Note that the ranges of the horizontal axes are different in each Figures 1 to 3. This reflects the fact that both tests are "consistent", and so their powers approach the value "1" (when the null becomes increasingly false) more rapidly as n grows.
Also, note that while the results in Figures 2 are similar to those seen in Table 1, those in Figure 3 are quite different. In the latter case the size-distortion for the t-test is upwards (at 7.8%) and its power curve lies above that of the permutation test. It's not that the t-test is (necessarily) more powerful than the the permutation test. We can't compare (true) power unless the two tests have the same significance level - and they don't in this case.
Figure 2
Figure 3
The detailed results from the Monte Carlo experiment on which Table 1 and Figures 2 and 3 are based can be found in an Excel spreadsheet on the data page for this blog.
Be careful!
Three caveats are in order here:
1. We set β1 = 1 in the DGP and this resulted in a mis-specified "fitted" regression model. This chosen value for β1 affects the (numerical) results. The magnitudes of the size distortions and the powers are specific to this choice.
2. We need to be careful, here, when we talk about comparisons between the powers of the two tests (and this comment is universally applicable). Strictly, power comparisons are valid when the tests have the same (actual, empirical) significance level. The only exception is when Test A has lower actual significance level than Test B, but Test A has a higher rejection rate than Test B when the null is false (i.e., higher "raw", or apparent, power") over the full parameter space. Then, Test A is more powerful than Test B.
In all other cases where there is size distortion, the "true" power will not be clear. One way to deal with this is to "size-adjust" any test that exhibits size distortion. This would be done, in our Monte Carlo experiment, by "jiggling" the critical values used for the t-test to ones that ensure that the test has a 5% rejection rate when the null is true. Then we could proceed to generate the corresponding power curves and make valid comparisons.
That's fine, but of course in practice we wouldn't know what modified critical values to use (unless we conducted a Monte Carlo experiment every time we undertook a real-life application). Perhaps this is worthy of another blog post at some stage, perhaps as an addition to my earlier ones on the basics of Monte Carlo simulation - here, here, and here.
3. As I stressed from the outset, model (1) is only a simple regression model. Multiple regression models are much more interesting, but this is where things get a lot trickier when it comes to constructing an exact permutation test. Quite a lot has been written about this problem. Some of the issues are discussed quite nicely by Kennedy (1995), but the (statistics) literature has moved along a bit since then and some of his suggestions are no longer supported. Some key references include Anderson and ter Braak (2003), Huh and Jhun (2001), Kim et al. (2000), LePage and Podgórski (1996), Oja (1987), and Schmoyer (1994).
Anderson and Robinson (2001) provide simulation evidence that favours the particular permutation procedure suggested by Freedman and Lane (1983). More recently, Nyblom (2015) introduces a permutation procedure based on the regression residuals, and illustrates its application to problems involving tests for autocorrelation, heteroskedasticity, and structural breaks.
Summing up1. We set β1 = 1 in the DGP and this resulted in a mis-specified "fitted" regression model. This chosen value for β1 affects the (numerical) results. The magnitudes of the size distortions and the powers are specific to this choice.
2. We need to be careful, here, when we talk about comparisons between the powers of the two tests (and this comment is universally applicable). Strictly, power comparisons are valid when the tests have the same (actual, empirical) significance level. The only exception is when Test A has lower actual significance level than Test B, but Test A has a higher rejection rate than Test B when the null is false (i.e., higher "raw", or apparent, power") over the full parameter space. Then, Test A is more powerful than Test B.
In all other cases where there is size distortion, the "true" power will not be clear. One way to deal with this is to "size-adjust" any test that exhibits size distortion. This would be done, in our Monte Carlo experiment, by "jiggling" the critical values used for the t-test to ones that ensure that the test has a 5% rejection rate when the null is true. Then we could proceed to generate the corresponding power curves and make valid comparisons.
That's fine, but of course in practice we wouldn't know what modified critical values to use (unless we conducted a Monte Carlo experiment every time we undertook a real-life application). Perhaps this is worthy of another blog post at some stage, perhaps as an addition to my earlier ones on the basics of Monte Carlo simulation - here, here, and here.
3. As I stressed from the outset, model (1) is only a simple regression model. Multiple regression models are much more interesting, but this is where things get a lot trickier when it comes to constructing an exact permutation test. Quite a lot has been written about this problem. Some of the issues are discussed quite nicely by Kennedy (1995), but the (statistics) literature has moved along a bit since then and some of his suggestions are no longer supported. Some key references include Anderson and ter Braak (2003), Huh and Jhun (2001), Kim et al. (2000), LePage and Podgórski (1996), Oja (1987), and Schmoyer (1994).
Anderson and Robinson (2001) provide simulation evidence that favours the particular permutation procedure suggested by Freedman and Lane (1983). More recently, Nyblom (2015) introduces a permutation procedure based on the regression residuals, and illustrates its application to problems involving tests for autocorrelation, heteroskedasticity, and structural breaks.
Here are the takeaways from this post:
- Permutation tests are nonparametric, or "distribution free". Unlike the usual tests that we use in econometric, we don't need to satisfy a whole lot of (possibly questionable) parametric and distributional assumptions.
- Permutation tests are easy to apply, even though they can be (moderately) computationally intensive.
- Permutation tests are "exact", in the sense that achieve precisely the significance level that we want them to. There is no "size distortion".
- Usually, we have lots of legitimate, and simple, choices for the test statistic in any given problem. In each case, the test will be exact, though its power may vary depending on our choice.
- A permutation test will still be exact, even if the model is mis-specified. This stands in stark contrast to the usual parametric tests that we use.
- A permutation test may be more, or less, powerful than its parametric counterparts, depending on the situation.
- If you plan to use permutation (randomization) tests in the context of the multiple regression model (or its extensions), then their are some pitfalls that you need to be aware of. Be sure to look at the references that I've supplied.
Other than that - Happy Testing!
References
Anderson, M. J. & J. Robinson, 2001. Permutation tests for linear models. Australian and New Zealand Journal of Statistics, 43, 75-88.
Anderson, M. J., & C. J. F. ter Braak, 2003. Permutation tests for multi-factorial analysis of variance. Journal of Statistical Computation and Simulation, 73, 85–113.
Edgington, E. S., 1987. Randomization Tests. Marcel Dekker, New York.
Freedman, D. & D. Lane, 1983. A nonstochastic interpretation of reported significance levels. Journal of Business and Economic Statistics, 1, 292-298.
Huh, M-H. & M. Jhun, 2001. Random permutation testing in multiple linear regression. Communications in Statistics - Theory Methods, 30, 2023–2032.
Kennedy, P. E., 1995. Randomization tests in econometrics. Journal of Business and Economic Statistics, 13, 85-94.
Kim, H.-J., M. P. Fay, E. J. Feuer, & D. Midthune, 2000. Permutation tests for jointpoint regression with applications to cancer rates. Statistics in Medicine, 19, 335–351.
LePage, R. & K. Podgórski, 1996. Resampling permutations in regression without second moments. Journal of Multivariate Analysis, 57, 119–141.
Noreen, E. W., 1989. Computer Intensive Methods for Testing Hypotheses: An Introduction. Wiley, New York.
Nyblom, J., 2015. Permutation tests in linear regression. Chapter 5 in K. Nordhausen & S. Taskinen (eds.), Modern Nonparametric, Robust and Multivariate Methods. Springer International, Switzerland.
Oja, H., 1987. On permutation tests in multiple regression and analysis of covariance problems. Australian Journal of Statistics, 29, 91–100.
Schmoyer, R. L., 1994. Permutation tests for correlation in regression errors. Journal of the American Statistical Association, 89, 1507–1516.



Thanks for this post. It would be interesting if you could discuss a little bit the comparison between this method and the bootstrap in this particular case (regression).
ReplyDeleteThe Bootstrap, of course involves sampling WITH replacement from a "population" generated using the observed test statistic. It's related, but quite different. I'll do a post on this before too long.
ReplyDeleteLovely post Prof. I will take time to look at the permutation test for split-plot design model.
ReplyDelete